Author Archives: Albert Bordons
We have good clostridia in the gut and some of them prevent allergies
21st March 2015
Clostridia: who are they ?
The clostridia or Clostridiales, with Clostridium and other related genera, are Gram-positive sporulating bacteria. They are obligate anaerobes, and belong to the taxonomic phylum Firmicutes. This phylum includes clostridia, the aerobic sporulating Bacillales (Bacillus, Listeria, Staphylococcus and others) and also the anaerobic aero-tolerant Lactobacillales (id est, lactic acid bacteria: Lactobacillus, Leuconostoc, Oenococcus, Pediococcus, Lactococcus, Streptococcus, etc.). All Firmicutes have regular shapes of rod or coccus, and they are the evolutionary branch of gram-positive bacteria with low G + C content in their DNA. The other branch of evolutionary bacteria are gram-positive Actinobacteria, of high G + C and irregular shapes, which include Streptomyces, Corynebacterium, Propionibacterium, and Bifidobacterium, among others.
Being anaerobes, the clostridia have a fermentative metabolism of both carbohydrates and amino acids, being primarily responsible for the anaerobic decomposition of proteins, known as putrefaction. They can live in many different habitats, but especially in soil and on decaying plant and animal material. As we will see below, they are also part of the human intestinal microbiota and of other vertebrates.
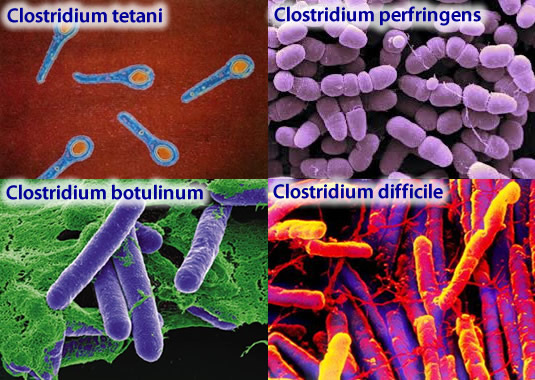
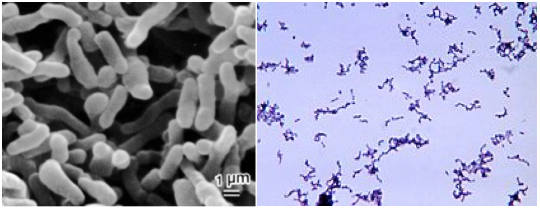
The best known clostridia are the bad ones (Figure 1): a) C. botulinum, which produces botulin, the botulism toxin, although nowadays has medical and cosmetic applications (Botox); b) C. perfringens, the agent of gangrene; c) C. tetani, which causes tetanus; and d) C. difficile, which is the cause of hospital diarrhea and some postantibiotics colitis.
Figure 1. The four more pathogen species of Clostridium. Image from http://www.tabletsmanual.com/wiki/read/botulism
Clostridia in gut microbiota
As I mentioned in a previous post (Bacteria in the gut …..) of this blog, we have a complex ecosystem in our gastrointestinal tract, and diverse depending on each person and age, with a total of 1014 microorganisms. Most of these are bacteria, besides some archaea methanogens (0.1%) and some eukaryotic (yeasts and filamentous fungi). When classical microbiological methods were carried out from samples of colon, isolates from some 400 microbial species were obtained, belonging especially to proteobacteria (including Enterobacteriaceae, such as E. coli), Firmicutes as Lactobacillus and some Clostridium, some Actinobacteria as Bifidobacterium, and also some Bacteroides. Among all these isolates, some have been recognized with positive effect on health and are used as probiotics, such as Lactobacillus and Bifidobacterium, which are considered GRAS (Generally Recognized As Safe).
But 10 years ago culture-independent molecular tools began to be used, by sequencing of ribosomal RNA genes, and they have revealed many more gut microorganisms, around 1000 species. As shown in Figure 2, taken from the good review of Rajilic-Stojanovic et al (2007), there are clearly two groups that have many more representatives than thought before: Bacteroides and Clostridiales.
Figure 2. Phylogenetic tree based on 16S rRNA gene sequences of various phylotypes found in the human gastrointestinal tract. The proportion of cultured or uncultured phylotypes for each group is represented by the colour from white (cultured) passing through grey to black (uncultured). For each phylogenetic group the number of different phylotypes is indicated (Rajilic-Stojanovic et al 2007)
In more recent studies related to diet such as Walker et al (2011) — a work done with faecal samples from volunteers –, population numbers of the various groups were estimated by quantitative PCR of 16S rRNA gene. The largest groups, with 30% each, were Bacteroides and clostridia. Among Clostridiales were included: Faecalibacterium prausnitzii (11%), Eubacterium rectale (7%) and Ruminococcus (6%). As we see the clostridial group includes many different genera besides the known Clostridium.
In fact, if we consider the population of each species present in the human gastrointestinal tract, the most abundant seems to be a clostridial: F. prausnitzii (Duncan et al 2013).
Benefits of some clostridia
These last years it has been discovered that clostridial genera of Faecalibacterium, Eubacterium, Roseburia and Anaerostipes (Duncan et al 2013) are those which contribute most to the production of short chain fatty acids (SCFA) in the colon. Clostridia ferment dietary carbohydrate that escape digestion producing SCFA, mainly acetate, propionate and butyrate, which are found in the stool (50-100 mM) and are absorbed in the intestine. Acetate is metabolized primarily by the peripheral tissues, propionate is gluconeogenic, and butyrate is the main energy source for the colonic epithelium. The SCFA become in total 10% of the energy obtained by the human host. Some of these clostridia as Eubacterium and Anaerostipes also use as a substrate the lactate produced by other bacteria such as Bifidobacterium and lactic acid bacteria, producing finally also the SCFA (Tiihonen et al 2010).
Clostridia of microbiota protect us against food allergen sensitization
This is the last found positive aspect of clostridia microbiota, that Stefka et al (2014) have shown in a recent excellent work. In administering allergens (“Ara h”) of peanut (Arachis hypogaea) to mice that had been treated with antibiotics or to mice without microbiota (Germ-free, sterile environment bred), these authors observed that there was a systemic allergic hyper reactivity with induction of specific immunoglobulins, id est., a sensitization.
In mice treated with antibiotics they observed a significant reduction in the number of bacterial microbiota (analysing the 16S rRNA gene) in the ileum and faeces, and also biodiversity was altered, so that the predominant Bacteroides and clostridia in normal conditions almost disappeared and instead lactobacilli were increased.
To view the role of these predominant groups in the microbiota, Stefka et al. colonized with Bacteroides and clostridia the gut of mice previously absent of microbiota. These animals are known as gnotobiotic, meaning animals where it is known exactly which types of microorganisms contain.
In this way, Stefka et al. have shown that selective colonization of gnotobiotic mice with clostridia confers protection against peanut allergens, which does not happen with Bacteroides. For colonization with clostridia, the authors used a spore suspension extracted from faecal samples of healthy mice and confirmed that the gene sequences of the extract corresponded to clostridial species.
So in effect, the mice colonized with clostridia had lower levels of allergen in the blood serum (Figure 3), had a lower content of immunoglobulins, there was no caecum inflammation, and body temperature was maintained. The mice treated with antibiotics which had presented the hyper allergic reaction when administered with antigens, also had a lower reaction when they were colonized with clostridia.
Figure 3. Levels of “Ara h” peanut allergen in serum after ingestion of peanuts in mice without microbiota (Germ-free), colonized with Bacteroides (B. uniformis) and colonized with clostridia. From Stefka et al (2014).
In addition, in this work, Stefka et al. have conducted a transcriptomic analysis with microarrays of the intestinal epithelium cells of mice and they have found that the genes producing the cytokine IL-22 are induced in animals colonized with clostridia, and that this cytokine reduces the allergen uptake by the epithelium and thus prevents its entry into the systemic circulation, contributing to the protection against hypersensitivity. All these mechanisms, reviewed by Cao et al (2014), can be seen in the diagram of Figure 4.
In conclusion, this study opens new perspectives to prevent food allergies by modulating the composition of the intestinal microbiota. So, adding these anti-inflammatory qualities to the production of butyrate and other SCFA, and the lactate consumption, we must start thinking about the use of clostridia for candidates as probiotics, in addition to the known Lactobacillus and Bifidobacterium.
Figure 4. Induction of clostridia on cytokine production by epithelial cells of the intestine, as well as the production of short chain fatty acids (SCFA) by clostridia (Cao et al 2014).
References
Cao S, Feehley TJ, Nagler CR (2014) The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS Lett 588, 4258-4266
Duncan SH, Flint HJ (2013) Probiotics and prebiotics and health in ageing populations. Maturitas 75, 44-50
Rajilic-Stojanovic M, Smidt H, de Vos WM (2007) Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 9, 2125-2136
Rosen M (2014) Gut bacteria may prevent food allergies. Science News 186, 7, 4 oct 2014
Russell SL, et al. (2012) Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 13(5):440–447
Stefka AT et al (2014) Commensal bacteria protect against food allergen sensitization. Proc Nat Acad Sci 111, 13145-13150
Tiihonen K, Ouwehand AC, Rautonen N (2010) Human intestinal microbiota and healthy aging. Ageing Research Reviews 9:107–16
Walker AW et al (2011) Dominant and diet-responsive groups of bacteria within the human colonic microbiota. The ISME J 5, 220-230
Bacteria in the gut are controlling what we eat
It seems to be so: the microbes in our gastrointestinal tract (GIT) influence our choice of food. No wonder: microbes, primarily bacteria, are present in significant amounts in GIT, more than 10 bacterial cells for each of our cells, a total of 1014 (The human body has about 1013 cells). This amounts to about 1-1.5 kg. And these bacteria have lived with us always, since all mammals have them. So, they have evolved with our ancestors and therefore they are well suited to our internal environment. Being our bodies their habitat, much the better if they can control what reaches the intestine. And how can they do? Then giving orders to the brain to eat such a thing or that other, appropriate for them, the microbes.
 Figure 1. “Command centre of the gastrointestinal tract” (own assembly, Albert Bordons)
Figure 1. “Command centre of the gastrointestinal tract” (own assembly, Albert Bordons)
Well, gone seriously, there is some previous work in this direction. It seems there is a relationship between preferences for a particular diet and microbial composition of GIT (Norris et al 2013). In fact, it is a two-way interaction, one of the many aspects of symbiotic mutualism between us and our microbiota (Dethlefsen et al 2007).
There is much evidence that diet influences the microbiota. One of the most striking examples is that African children fed almost exclusively in sorghum have more cellulolytic microbes than other children (De Filippo et al 2010).
The brain can also indirectly influence the gut microbiota by changes in intestinal motility, secretion and permeability, or directly releasing specific molecules to the gut digestive lumen from the sub epithelial cells (neurons or from the immune system) (Rhee et al 2009).
The GIT is a complex ecosystem where different species of bacteria and other microorganisms must compete and cooperate among themselves and with the host cells. The food ingested by the host (human or other mammal) is an important factor in the continuous selection of these microbes and the nature of food is often determined by the preferences of the host. Those bacteria that are able to manipulate these preferences will have advantages over those that are not (Norris et al 2013).
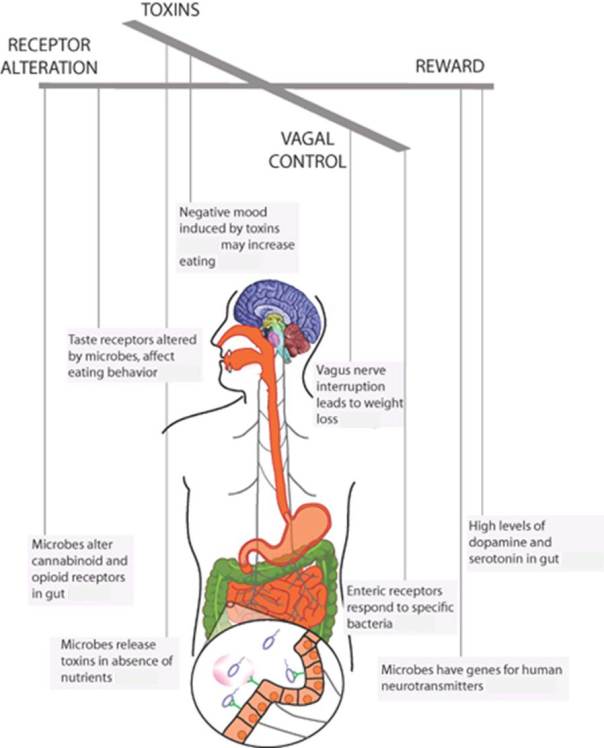
Recently Alcock et al (2014) have reviewed the evidences of all this. Microbes can manipulate the feeding behaviour of the host in their own benefit through various possible strategies. We’ll see some examples in relation to the scheme of Figure 2.
Figure 2. As if microbes were puppeteers and we humans were the puppets, microbes can control what we eat by a number of marked mechanisms. Adapted from Alcock et al 2014.
People who have “desires” of chocolate have different microbial metabolites in urine from people indifferent to chocolate, despite having the same diet.
Dysphoria, id est, human discomfort until we eat food which improve microbial “welfare”, may be due to the expression of bacterial virulence genes and perception of pain by the host. This is because the production of toxins is often triggered by a low concentration of nutrients limiting growth. The detection of sugars and other nutrients regulates virulence and growth of various microbes. These directly injure the intestinal epithelium when nutrients are absent. According to this hypothesis, it has been shown that bacterial virulence proteins activate pain receptors. It has been shown that fasting in mice increases the perception of pain by a mechanism of vagal nerve.
Microbes can also alter food preferences of guests changing the expression of taste receptors on the host. In this sense, for instance germ-free mice prefer more sweet food and have a greater number of sweet receptors on the tongue and intestine that mice with a normal microbiota.
The feeding behaviour of the host can also be manipulated by microbes through the nervous system, through the vagus nerve, which connects the 100 million neurons of the enteric nervous system from the gut to the brain via the medulla. Enteric nerves have receptors that react to the presence of certain bacteria and bacterial metabolites such as short chain fatty acids. The vagus nerve regulates eating behaviour and body weight. It has been seen that the activity of the vagus nerve of rats stimulated with norepinephrine causes that they keep eating despite being satiated. This suggests that GIT microbes produce neurotransmitters that can contribute to overeating.
Neurotransmitters produced by microbes are analogue compounds to mammalian hormones related to mood and behaviour. More than 50% of dopamine and most of serotonin in the body have an intestinal origin. Many persistent and transient inhabitants of the gut, including E. coli, several Bacillus, Staphylococcus and Proteus secrete dopamine. In Table 1 we can see the various neurotransmitters produced by GIT microbes. At the same time, it is known that host enzymes such as amine oxidase can degrade neurotransmitters produced by microorganisms, which demonstrates the evolutionary interactions between microbes and hosts.
Table 1. Diversity of neurotransmitters isolated from several microbial species (Roschchina 2010)
| Neurotransmitter | Genera |
| GABA (gamma-amino-butyric acid) | Lactobacillus, Bifidobacterium |
| Norepinephrine | Escherichia, Bacillus, Saccharomyces |
| Serotonin | Candida, Streptococcus, Escherichia, Enterococcus |
| Dopamine | Bacillus, Serratia |
| Acetylcholine | Lactobacillus |
Some bacteria induce hosts to provide their favourite nutrients. For example, Bacteroides thetaiotaomicron inhabits the intestinal mucus, where it feeds on oligosaccharides secreted by goblet cells of the intestine, and this bacterium induces its host mammal to increase the secretion of these oligosaccharides. Instead, Faecalibacterium prausnitzii, a not degrading mucus, which is associated with B. thetaiotaomicron, inhibits the mucus production. Therefore, this is an ecosystem with multiple agents that interact with each other and with the host.
As microbiota is easily manipulated by prebiotics, probiotics, antibiotics, faecal transplants, and changes in diet, controlling and altering our microbiota provides a viable method to the otherwise insoluble problems of obesity and poor diet.
References
Alcock J, Maley CC, Aktipis CA (2014) Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. BioEssays 36, DOI: 10.1002/bies.201400071
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, et al (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107:14691–6
Dethlefsen L, McFall-Ngai M, Relman DA (2007) An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811-818
Lyte M (2011) Probiotics function mechanistically as delivery for neuroactive compounds: Microbial endocrinology in teh design and use of probiotics. BioEssays 33:574-581
Norris V, Molina F, Gewirtz AT (2013) Hypothesis: bacteria control host appetites. J Bacteriol 195:411–416
Rhee SH, Pothoulakis C, Mayer EA (2009) Principles and clinical implications of the brain–gut–enteric microbiota axis. Nature Reviews Gastroenterology and Hepatology 6:306-314
Roschchina VV (2010) Evolutionary considerations of neurotransmitters in microbial, plant, and animal cells. In Lyte M, Freestone PPE, eds; Microbial Endocrinology: Interkingdom Signaling in Infectious Disease and Health. New York: Springer. pp. 17–52
Surprising: bacteria of human acne passed to the vineyard !!
It is really surprising, but it seems so: Italian and Austrian researchers have published a paper (Campisano et al. 2014) which shows that the bacterial species Propionibacterium acnes, related to human acne, can be found as obligate endophytes in bark tissues of Vitis vinifera, the grapevine.
Some bacterial pathogens of humans, such as Salmonella, are able to colonize plant tissues but temporarily and opportunistically (Tyler & Triplett 2008). In fact, there is a temporary mutual benefit between plants and bacteria, so some of these enterobacteria pathogenic to plants do not live endophytically and can be beneficial for them. These pathogens to humans, in its life cycle, use plants as alternative hosts to survive the environment, passing to the plants through contaminated irrigation water. Therefore, some bacteria are often temporary endophyte guests of plants.
But on the other hand, there are relatively rare cases of bacteria changing the host and adapting to the new host, finally being endophytes. This horizontal transfer happens mostly between evolutionarily close hosts, such as symbiotic bacteria of aphids (insects), which has proven to transfer to other species of aphids (Russell & Moran 2005). It has also been suggested the horizontal transfer of beneficial lactic acid bacteria (Lactobacillus reuteri) in the intestinal tract of vertebrates, since strains of this L. reuteri are similar in several species of mammals and birds.
Well, going beyond, the work of Campisano et al. subject of this review, concludes that bacteria associated with human acne should have passed on the vine, that is, the bacteria would have made a horizontal transfer interregnum, from plants to mammals.
Propionibacterium acnes type Zappae
Acne, as you know, is a common human skin disease, consisting of an excess secretion of the pilosebaceous glands caused by hormonal changes, especially teenagers. The glands become inflamed, the pores obstructed and scarring appears. The microorganism associated with these infections is the opportunistic commensal bacterium P. acnes, a gram-positive anaerobic aero tolerant rod, which fed fatty acids produced by the glands.
Young with acne (Wikimedia, public)
Propionibacterium acnes at the scanning electron microscope (left) and dyed with violet crystal (right). From Abate ME (2013) Student Pulse 5, 9, 1-4.
Interestingly, other species of the same genus Propionibacterium well known in microbial biotechnology industry are used for the production of propionic acid, vitamin B12, and the Swiss cheeses Gruyere or Emmental.
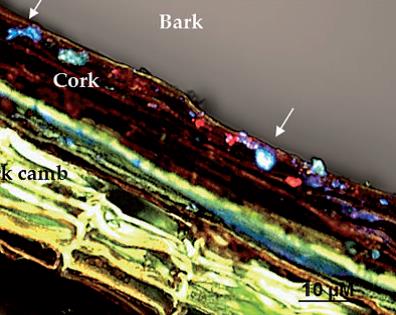
Campisano et al. have made a study of the vineyard endomicrobioma by the sequencing technique (Roche 454) amplifying the V5-V9 hyper variable region of the bacterial 16S rDNA present in the tissues of vine. In 54 of the 60 plants analyzed, between 0.5% and 5% of the found sequences correspond to the species Propionibacterium acnes. This observation has been confirmed by fluorescent in situ hybridization (FISH) with fluorochromes and specific probes of P. acnes.
Location of P. acnes (fluorescent blue spots) in the bark of a vine stem, seen with FISH microscopy with specific probes for this bacterium (Campisano et al 2004).
The authors of this work proposed for this bacterium the name of P. acnes Zappae, in memory of the eccentric musician and composer Frank Zappa, to emphasize the unexpected and unconventional habitat of this type of P. acnes.
Frank Zappa (1940-1993), the eccentric and satiric singer, musician and composer. Photo: Frank Zappa reviews.
And how did this human bacteria arrive into the vineyard?
To solve this riddle, Campisano et al. have taken the 16S rDNA sequences and from other genes (recA and tly) from these strains of P. acnes Zappae found in vine and have compared with those P. acnes of human origin in databases. Comparing phylogenies and clusters deducted from them, these researchers have concluded that P. a. Zappae has diversified evolutionarily recently. Studying in detail the recA gene sequences of P. a. Zappae, and taking into account the likely mutation rate and generation time (about 5 hours), they deduce that the diversification from other P. acnes occurred 6000-7000 years ago.
This date coincides with the known domestication of the vine by humans, which is believed to have occurred about 7000 years ago in the southern Caucasus, between the Black Sea and the Caspian Sea, the area of modern Turkey, Georgia, Armenia and Iran (Berkowitz 1996). The vineyard has its origins in a wild subspecies of Vitis that survived the Ice Age and was domesticated. This plant came out to three subspecies, and one of them, Vitis vinifera pontica, spread in the mentioned area and further south in Mesopotamia and then to all south Europe thanks to the Phoenicians.
Therefore, the conclusion is that P. acnes Zappae originated from human P. acnes 7000 years ago, by contact of human hands with grapes and other parts of the vineyard during the harvest and carrying them. As the authors say, this case would be the first evidence of horizontal transfer interregnum, from humans to plants, of a obligate symbiotic bacterium. This also makes more remarkable the adaptability of bacteria. Their ability to exploit new habitats can have unforeseen impacts on the evolution of host-symbiont relationship or even host-pathogen.
Harvesting by hand in Chile (Fine Wine and Good Spirits)
References
Berkowitz M (1996) World’s earliest wine. Archaeology 49, 5, Sept./Oct.
Campisano Aet al. (2014) Interkingdom transfer of the acne-causing agent, Propionibacterium acnes, from human to grapevine. Mol Biol Evol 31, 1059-1065.
Gruber K (4 march 2014) How grapevines got acne bacteria. Nature News 4 march 2014.
Russell JA, NA Moran (2005) Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl Environ Microbiol 71, 7987-7994.
Tyler HL, EW Triplett (2008) Plants as a habitat for beneficial and/or human pathogenic bacteria. Ann Rev Phytopathol 46, 53-73.
Wikipedia, of course: Propionibacterium acnes, Vitis, …
Walter J, RA Britton, S Roos (2011) PNAS 108, 4645-4652.
Synthesis of amino acids by impacts of comets, and other things related with life’s origin
A few weeks ago I saw the following headline in “Recercat” (the electronic newsletter of research in Catalonia): “Científics descobreixen com es formen les molècules bàsiques de la vida“, that is, “Scientists discover how the basic molecules of life are made”. Oops what a heading ! I was very surprised, of course, because I have always been passionate for this subject of life’s origin. So, I quickly read it, and looked for the original article .
Well, once seen in detail, it is clear that the heading is very, very exaggerated, as supposed at first glance. Before discussing why I think it is exaggerated, I want to mention another detail of this headline that I think is superflous: Why the word “scientists” ? Whoelse might be working on the study of the formation of the basic molecules of life besides scientists ? Politicians perhaps? or economists or maybe the bishops ? It is clear that must be scientists and therefore it is no need to say it. The heading would be enough like this: “It has been discovered how…. ” . Or else, it could tell where scientists are from: “English scientists, or American, or Japanese, have discovered … “.
Well, going to the specific discovery, and as Recercat itself summarizes, some Americans and British researchers (Martins et al. 2013) have published in Nature Geoscience their laboratory work, where they simulated the impact of comets on the surface of a planet, shooting a projectile with a compressed air pistol at speeds of 7 km/s (25,000 km/h). They have seen that by the impact and the heat generated, amino acids are synthesized from water, CO2 and ammonia. This is what they call “shock synthesis”. Among others, the amino acids detected were glycine, D-alanine, L-alanine, amino isobutyric acid, isovaline, norvaline and other precursors of amino acids, both isomers D and L. The amounts detected were between nanograms and one microgram.
This synthesis process shows that a simple mechanism, such as the impact of comets on the surface of a rocky planet, can transform basic inorganic molecules to more complex organic molecules such as amino acids, which are the monomers of proteins, basic constituents of all living beings. And therefore, this shock synthesis could have been a step in the appearance of terrestrial life.
Now, is this the first work to demonstrate the possible formation of basic molecules of life ? Well, absolutely not ! This work has its value but does not deserve this headline so exaggerated.
So, let’s briefly review what is already known, since more than 50 years, many different researchers have worked on this topic and have been discovering aspects that reinforce the scientific hypotheses of abiogenesis. This, also known as biopoesis, is the natural process by which living organisms originated on earth from simple molecules about 3,700 million years ago.
Biopoesis, the process of emergence of living beings on Earth
This process of biopoesis necessarily involved several steps:
1) The formation and appearance of basic organic molecules of living beings, it is, the monomers such as amino acids, monosaccharides, fatty acids and nitrogenous bases.
2) From the above monomers, the formation of biogenic macromolecules, it is, polysaccharides, polypeptides, lipoids and others, by polymerization, probably on the surface of inorganic substrates such as clay or iron minerals.
3) And the formation of the protobionts, the precursors of first cells, from macromolecules. This key step, the most difficult to prove, was probably linked to the parallel acquisition of the three basic properties of living beings: i) a structure envelope (the membrane) of lipidic consistency; ii) some reactions transforming nutrients and energy (rudimentary metabolism); and iii) the ability of transfering characteristics to the offspring (hereditary mechanism) with an information-carrying molecule, probably RNA.
By the moment I will leave the steps 2 and 3, maybe for discussing in a future post, and I will focus on the first one, related to the mentioned article of Martins et al. (2013). This formation and appearance of compounds in the early Earth may have been by three mechanisms: a) in situ production, b) contributions from abroad: c) synthesis due to impacts.
These three categories of mechanisms were already raised as an inventory of the possible origins of life in 1992 in an article of Nature by Christopher Chyba and Carl Sagan, the famous pioneer of exobiology and science writer, very known by the extraordinary TV series Cosmos, and author of the phrase “We are stardust“. Coincidentally, he was the first husband of Lynn Margulis, who spread the endosymbiotic theory of the bacterial origin of mitochondria and chloroplasts.
Endogenous synthesis of organic compounds on the primitive Earth
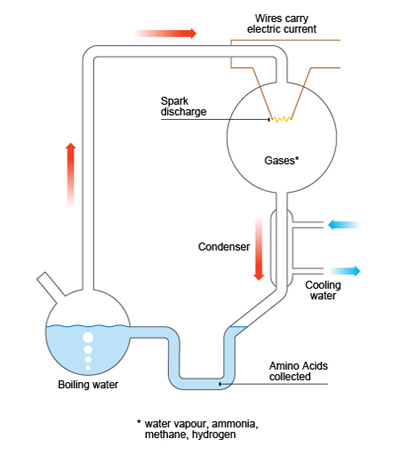
Well, as I said, it has been demonstrating since many years the possibility of the formation of organic molecules in situ, id est, endogenous synthesis in the primitive Earth, without external inputs. The Oparin’s hypothesis that the reducing anaerobic conditions of the early atmosphere, along with solar energy, should favoured the synthesis of organic molecules forming the “prebiotic soup”, was demonstrated as possible by the known experiments of Miller and Urey:
In 1952, the graduate student Stanley Miller with his professor Harold Urey introduced a mixture of water, hydrogen, methane and ammonia in a cyclical container, where electrical sparks were applied. A week later, when analyzing the components, they found that 15% of the carbon coming from methane had been transformed in various organic compounds, including 5 amino acids, both D-and L-.
Schema of the experiments of Miller and Urey. From GCSE-Bitesize (BBC).
Recently (Parker et al 2011) the tubes with the extracts of original Miller- Urey experiments have been analyzed again with current sophisticated analytical techniques and equipment, and many more compounds that the found originally in the 1950s have been discovered, in particular 23 other amino acids.
The synthesis of these organic molecules in the early Earth was probably facilitated by energy sources of the atmospheric activity such as electrical discharges that were used by Miller and Urey experiments, but there were other possibilities, such as the sunlight, with more UV radiation than at present (there was no ozone layer since it was formed later, from oxygen), and also more volcanic activity and radioactivity in a younger Earth, and impacts of meteorites and comets, which is related to the mentioned work of Martins et al. (2013).
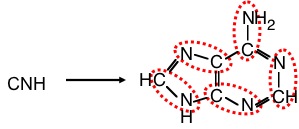
Another important contribution in the search for prebiotic organic synthesis was the demonstration made by Joan Oró (born in Lleida, Catalonia) working at NASA (Oró 1961) that adenine can be synthesized heating ammonium cyanide solutions. Similarly, recently it has been demonstrated the synthesis of pyrimidines (cytosine and uracil), adenine and triazines (other nitrogenous bases) from urea by freeze-thaw cycles and electric shocks (Menor-Salván et al. 2007).
As demonstrated by Joan Oró, adenine can be synthesized from 5 molecules of hydrogen cyanide. Adenine is a key molecule for life, since it is part of nucleic acids and ATP.
Contribution of organic molecules by extraterrestrial objects
In addition to the in situ synthesis of organic compounds in the primitive Earth, they could also have come from outside. The contribution of organic molecules by extraterrestrial objects, comets or meteorites or other, it is increasingly more evident scientifically. Recent studies suggest that the so-called massive bombing that took place 3.5 billion years ago provided an amount of organic compounds comparable to those produced in situ.
Simulation of meteor shower similar to this of the early Earth. From AZ-Revista de Educación y Cultura
It has been shown that organic compounds are relatively common in extraterrestrial space, especially in the outer solar system where volatile compounds are not evaporated by solar heat. Many comets have an outer layer of a material with appearance of tar, which contains organic compounds formed by reactions caused by radiation, especially UV. Apart from that long ago they had been detected by telescope spectrography, a few years ago it has been identified in situ the amino acid glycine in comet Wild -2 in samples taken by NASA’s Stardust spacecraft (Dolmetsch 2006) .
The Murchison meteorite, about 100 kg, fell in Australia in 1969 and broke in several fragments that have been well studied. This meteorite is of type carbonaceous chondrites, which are rich in carbon, and indeed contains amino acids, both common (glycine, alanine and glutamic acid) as the most unusual (isovaline, pseudoleucine), with concentrations up to 60 ppm (Kvenvolden et al. 1970). It also contains aromatic and aliphatic hydrocarbons, alcohols and other organic compounds such as carboxylic acids and fullerenes.
A fragment of the Murchison meteorite, fell in Australia in 1969, of the type carbonaceous chondrites, which contains amino acids and other organic compounds. Image from Wikipedia
The isotope ratio 12C/13C of uracil and other organic compounds in the Murchison indicates an origin no terrestrial (Martins et al. 2008). In fact, besides the Murchison, the analyses made with many other meteorites show that organic compounds can be formed in outer space.
Model studies done with computer suggest that prebiogenic organic compounds might be formed in the protoplanetary disk of dust that surrounded the sun before the formation of the Earth, and that the same process may happen around other stars (Moskowitz 2012).
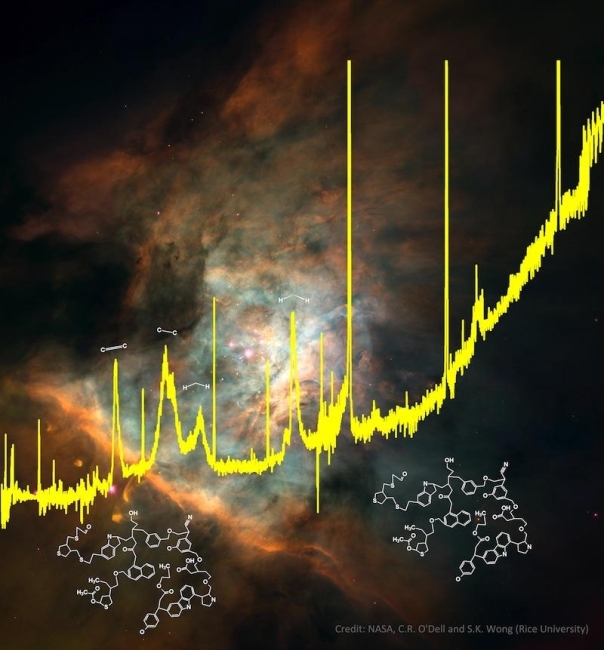
Moreover, studies of infrared emission spectra (Kwok & Zhang 2011) of cosmic dust have concluded that complex organic molecules are produced in the supernova stars, and that these molecules are expelled to the interstar space by effect of the explosion of the star. Surprisingly, this organic dust is similar to compounds found in meteorites. Since meteorites are remnants of the early solar system, we can now suggest that organic compounds found in meteorites had formed in distant stars .
Infrared spectrum of organic compounds, superimposed on an image of the Orion Nebula where these complex organic compounds (with the formulas) have been found. Image taken from NASA (C.R. O’Dell and S.K. Wong, Rice University.
In recent years there has been a breakthrough in the detection of organic molecules in galactic space thanks to the radio telescopes such as the Green Bank (100 m diameter) in West Virginia (USA), and the ALMA (Atacama Large Millimeter Array, at 5000 m in the Atacama Desert, northern Chile) which is composed of 66 radio antennas of 12 m diameter connected to each other by optical fiber. These astronomical radio telescopes or interferometers capture wavelengths around mm.
With these telescopes, as commented by Pere Brunet last month in his post post “Som pols d’estrelles ?” (Are we stardust ?, of the blog Fractal Ara-Ciència), different compounds have been detected around other stars: propenal, cyclo-propenone, acetamide, and glycolaldehyde (CHO-CH2OH). The latter is quite significant, since it is the smallest sugar and it is necessary for the formation of RNA. It has been detected with ALMA, around a young binary sun-type star (IRAS 16293-2422 ), 400 light years from Earth, relatively close, within the Milky Way (Jørgensen et al. 2012) .
Synthesis of organic compounds due to impacts of extraterrestrial objects
Finally, we have this third mechanism related with the article object of this post (Martins et al. 2013). Well, as we said before, this possibility was already reviewed by Chyba & Sagan (1992), because experiments in this direction had been made years ago. Carl Sagan itself, with other authors (Bar-Nun et al. 1970) had shown that applying a thermal shock, simulating impacts of comets and micrometeorites, to a gas mixture similar to the primitive atmosphere, there appeared amino acids.
However, thinking about the impacts of extraterrestrial bodies the first idea is the opposite, that they are antagonists of life on Earth because we remember the impact which caused extinctions and catastrophes such as the asteroid 65 million years falling in the Chicxulub crater in Yucatan, or a much smaller scale, the meteorite that fell last year in Chelyabinsk, Russia, with the appearance of fireballs. However, although these collisions can cause adverse effects on living beings where they fall, at the same time the energy released by the shock can be a source of reactions that generate prebiogenic organic compounds, as evidenced by the work of Martins et al. (2013).
At the same time, it has been shown that biological compounds that were present in the meteorite can “survive” impacts. Indeed, it has been shown that these compounds may be captured in the carbonaceous pores, inside the molten material due to the temperature and pressure of impact, particularly in analyses made with material of Darwin Crater in Tasmania, coming from a meteorite that impacted 800,000 years ago (Howard et al. 2013).
Besides from Martins et al., other studies have also simulated the effects of impacts. Furukawa et al. (2009) simulated the impact of a meteorite chondrite-type in a primitive ocean. They used a propulsion gun to create a high-speed impact in a mixture of carbon, iron, nickel, nitrogen and water, and then they recovered several organic molecules, including fatty acids, amines and one amino acid.
Thus, these experiments suggest that the impact of frequent extraterrestrial bodies in the primitive Earth had resulted in a great contribution to the formation of many different organic compounds, and as I said, adding that to the contribution of the already previously synthesized in outer space and to in situ synthesis in the same Earth.
Bibliography
Bar-Nun A, Bar-Nun N, Bauer SH, Sagan C. 1970. Shock synthesis of amino acids in simulated primitive environments. Science 168, 470-472.
Chyba C, Sagan C. 1992. Endogenous production, exogenous delivery and impact-shock synthesis of organic molecules: an inventory for the origins of life. Nature 355, 125–32
Dolmetsch C. 2006. NASA Spacecraft Returns With Comet Samples After 2.9 Bln Miles. Bloomberg.com. 2006-01-15
Editorial. 2013. The upside of impacts. Nature Geoscience 6, 987.
Furukawa Y, Sekine T, Oba M, Kakegawa T, Nakazawa H. 2009. Biomolecule formation by oceanic impacts on early Earth. Nature Geoscience 2, 62–66
Generalitat de Catalunya. 2014. “Científics descobreixen com es formen les molècules bàsiques de la vida”. Recercat 94, gener 2014.
Howard KT et 12 al. 2013. Biomass preservation in impact melt ejecta. Nature Geoscience 6, 1018-1023.
Jørgensen JK, Favre C, Bisschop SE, Bourke TL, van Dishoeck EF, Schmalzl M. 2012. Detection of the simplest sugar, glycolaldehyde, in a solar-type protostar with ALMA. Astrophysical Journal Letters 757, L4, 1-13.
Kvenvolden KA, Lawless J, Pering K, Peterson E, Flores J, Ponnamperuma C, Kaplan IR, Moore C. 1970. Evidence for extraterrestrial amino-acids and hydrocarbons in the Murchison meteorite. Nature 228, 923–926
Kwok S, Zhang Y. 2011. Mixed aromatic–aliphatic organic nanoparticles as carriers of unidentified infrared emission features. Nature, DOI:10.1038/nature10542
Martins Z, Botta O, Fogel ML, Sephton MA, Glavin DP, Watson JS, Dworkin JP, Schwartz AW, Ehrenfreund P. 2008. Extraterrestrial nucleobases in the Murchison meteorite. Earth and Planetary Science Letters 270, 130–136
Martins Z, MC Price, N Goldman, MA Sephton, MJ Burchell. 2013. Shock synthesis of amino acids from impacting cometary and icy planet surface analogues. Nature Geoscience 6, 1045-1049.
Moskowitz C. 2012. Life’s Building Blocks May Have Formed in Dust Around Young Sun. Space.com
Menor-Salván C, Ruiz-Bermejo DM, Guzmán MI, Osuna-Esteban S, Veintemillas-Verdaguer S. 2007. Synthesis of pyrimidines and triazines in ice: implications for the prebiotic chemistry of nucleobases. Chemistry 15, 4411–8.
Oparin A. 1952. The origin of life. New York: Dover.
Oró J. 1961. Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive Earth conditions. Nature 191, 1193–4.
Parker ET, Cleaves HJ, Dworkin JP et al. 2011. Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. PNAS 108, 5526–31.
Wikipedia: http://en.wikipedia.org/wiki/Abiogenesis (very good review of life’s origin andthe different hypothesis)
An “habitable” lake in Mars 3800 million years ago
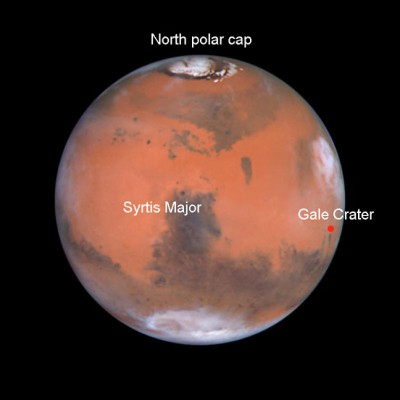
As I mentioned in this blog a year and a half before (August 30th, 2012 “Is there life on Mars?”), the Curiosity rover began to walk by Mars, specifically inside the Gale crater, carrying its sophisticated instruments to analyze rocks, soil and atmosphere, that is, a well equipped geochemical laboratory analyzing the surface of another planet for the first time. You can see a video of 2 minutes of the 1st year of Curiosity here. As you know, one of the objectives of this analysis is to find evidence of whether there were favorable conditions for life on Mars, albeit in very bygone eras.
The Curiosity rover is walking inside Gale Crater, east of Syrtis Major, the Mars most prominence visible through a telescope. Image NASA/ESA/Hubble.
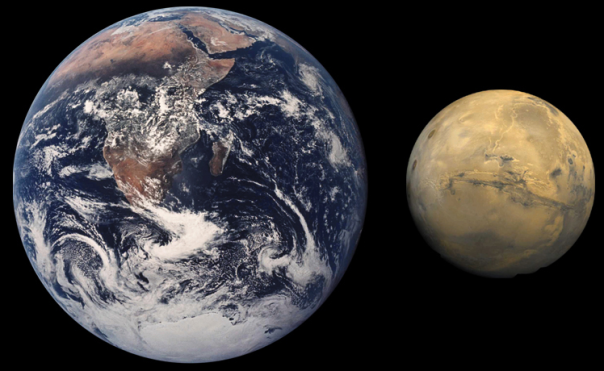
Mars is a planet with some similar features on Earth, such as the length of day, slightly more than 24 hours, but the greater distance from the Sun and very tenuous atmosphere make the average temperature to be -63 ° C, with daily oscillations from +20°C to -140°C. Having no oceans, the land surface of Mars is equivalent to the Earth, because Mars is smaller.
Comparison of Earth and Mars. The diameter of Mars is 3400 km, approx half of the Earth’s, but excluding oceans, the solid surface is similar on both planets. Image taken from T.E. Harrison, New Mexico State University.
Despite the difficulties to demonstrate the current existence of liquid water on Mars, from some years ago evidences have been found that there was water in the Martian surface, and it is being confirmed that during the first thousand million years there was plenty of water on Mars (Remind that both Mars and Earth are about 4.5 billion years).
Appearance of a region of Mars indicating the flow of water in the distant past. Image taken from T.E. Harrison, New Mexico State University.
Water history in Mars, in billions of years. Image from T.E. Harrison, New Mexico State University.
Curiosity shows an “habitable” lake 3800 million years ago
Thus, Curiosity rover has been taking samples in various ways (see in Figure below the 5 cm holes taking samples) and analyzing them. Studying the data, scientists have been able to write a series of articles that have been published in Science and other prestigious journals.
Two 5 cm holes made by Curiosity rover sampling in Yellowknife Bay of Gale Crater on Mars (NASA/JPL-Caltech/MSSS)
Among other articles, by last May was already published one (Williams et al 2013) with observations that the material extracted from the Yellowknife Bay of Gale Crater shows textures typical of fluvial sedimentary conglomerates, with rounded pebbles that indicate fluvial abrasion, and by their characteristics a water flow of nearly 1 meter per second has been deduced, with a depth of near 1 meter. Therefore, when this area was formed the climatic conditions, with abundant rivers, would have been very different from the current hyperarid and very cold environment.
A few weeks ago, on December 9th, Science Online has released a special edition dedicated to the latest findings of Curiosity, with 6 articles asserting that at Gale Crater there would have been a lake theoretically habitable for some organisms.
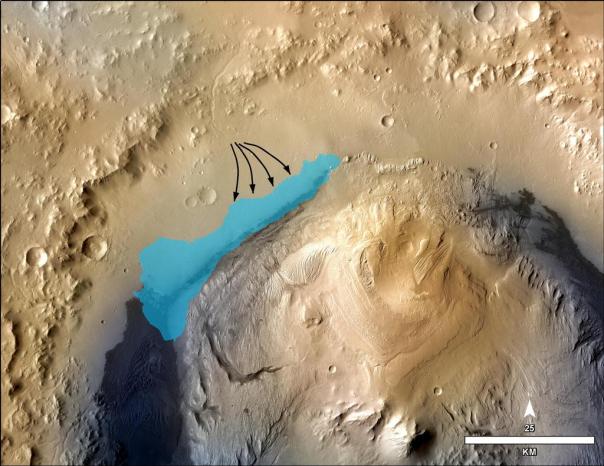
In one of the articles, maybe the most relevant one, Grotzinger et al (2013) describe the sedimentary rocks found by the rover and demonstrate that it corresponded to an aqueous environment with neutral pH, low salinity and different redox states of Fe and S. The presence of C, H, O, S, N and P (i.e., all biogenic elements) is also shown, and the authors estimate that this favorable environment for life could have lasted some few hundred thousand years, and therefore it demonstrates the biological feasibility of this fluvio-lacustrine environment of post-Noachian Mars, that is, about 3800 million years ago.
Therefore, these scientists deduced that this lake had fresh water, a water we could drink, and that around this lake various fluvial and lacustrine environments were present. This environment should be habitable for some organisms, and very similar to some current earth environments. The commented chemical compounds would be suitable for the survival of bacteria chemolithotrophic bacteria, id est, those which use inorganic compounds (such as Fe or S) as an energy source and use CO2 as a carbon source. On Earth we have many kind of these microbes, such as iron bacteria (Thiobacillus) or sulfur bacteria (including some sulfur thermophilic archaea) and the nitrifying ones. Although most of these chemolithotrophic bacteria on Earth are aerobic because they use atmospheric oxygen as electron acceptor, this metabolism is also known in some anaerobes, such as those that do the anoxic ammonium oxidation (process “Anammox”), for example Brocadia anammoxidans, or the same nitrifying bacteria in anaerobic conditions.
Therefore, with this habitable lake for some time, and with other similar environments, we can not reject the possibility that living beings had been sometimes in Mars.
View from Yellowknife Bay in Gale Crater, looking W-NW. This area of sedimentary deposits was the bottom of a freshwater lake. Photo: NASA/JPL-Caltech.
Estimated size and shape of the lake that was in the Gale crater. There must have been other similar. The arrows indicate the direction of the alluvial fan that flowed into the crater from its wall. Photo: NASA/JPL-Caltech/MSSS.
Another aspect studied in this extra issue of Science is the possible presence of methane in the Martian atmosphere (Webster et al 2013). Just like on Earth, methane is a potential sign of biological activity in the past. Previous observations made from Earth or from the Mars orbit speculated on the presence of some 10 ppb of methane, and its subsurface biological origin was also discussed. Nevertheless, in the measurements made in situ by laser spectrometry of Curiosity rover using a specific spectral methane pattern, it has not been detected since the values found are lower than 1 ppb. This reduces the probability of recent methanogenic microbial activity on Mars and it is limiting the possible contribution of recent geological and extraplanetary sources.
Finally, in another article (Hassler et al 2013) about measures of cosmic rays on the surface of Mars that Curiosity has made over a year, models of radiation are elaborated, for the time calculation of possible subsurface microbial survival, and also for the preservation of organic compounds of biological origin finding them billions of years later. Unfortunately, the conclusion is that the powerful effect of cosmic rays would cause very difficult any of these possibilities. Unlike the Earth, the magnetic field of Mars is very weak and the atmosphere too, things that help bombardment of cosmic rays and the solar wind on the Martian surface. However, there is evidence that in the past Mars had a magnetic field more effective, so hope is not lost …..
Bibliography
Achenbach J. 2013. NASA Curiosity rover discovers evidence of freshwater Mars lake. The Washington Post, Health & Science online Dec 9.
Anderson PS. 2013. Curiosity rover confirms ancient martian lake. Spaceflight Insider online Dec 11.
Grotzinger JP et 72 al. 2013. A Habitable Fluvio-Lacustrine Environment at Yellowknife Bay, Gale Crater, Mars. Science DOI: 10.1126/science.1242777 online Dec 9, 2013.
Harrison, T.E., New Mexico State University: http://astronomy.nmsu.edu/tharriso/ast105/Mars.html
Hassler DM et 23 al. 2013. Mars’ Surface Radiation Environment Measured with the Mars Science Laboratory’s Curiosity Rover. Science DOI: 10.1126/science.1244797 online Dec 9, 2013.
Kerr, RA. 2013. New Results Send Mars Rover on a Quest for Ancient Life. Science Now News, online Dec 9, 2013.
Science Special Collection Curiosity 2013. Curiosity at Yellowknife Bay, Gale Crater. Online: http://www.sciencemag.org/site/extra/curiosity/
Webster CR et al 2013. Low upper limit to methane abundance on Mars. Science 18 October 2013, Vol. 342 no. 6156 pp. 355-357
Williams RME et 38 al. 2013. Martian Fluvial Conglomerates at Gale Crater. Science 31 May 2013, Vol. 340 no. 6136 pp. 1068-1072
Agromicrobiome: microorganisms from the roots of crop plants
All we that studied “Bios” probably remember two known aspects of the symbiotic relationships of plant roots with microorganisms:
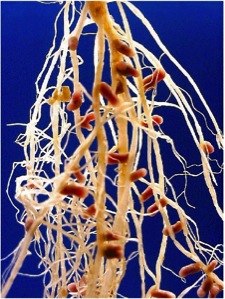
1) The bacterial Rhizobium nodules on the roots of legumes (Figure 1). These bacteria, with the nitrogenase complex, are among the few organisms capable of fixing atmospheric N2 transforming it into organic nitrogen, which is used by the plant, and symbiotically, the plant provides organic compounds to the bacteria. Thanks to these bacteria, plants such as legumes do not require nitrogen fertilizers.
Figure 1. Rhizobium nodules
2) The mycorrhizae, that is, the symbiotic relationships between fungi and plant roots. The most commonly known are the mushrooms always associated with some trees (Figure 2), such as the Lactarius sanguifluus associated with pines. In fact, mycorrhizae are present in most plants. Through this symbiosis, the fungi receive organic nutrients of the plant, and this can capture more easily water and mineral nutrients (especially P, Zn and Cu) by means of the fungus. In addition, mycorrhizae increase the resistance of plants to diseases coming from the soil and facilitate them inhabiting badlands.
Figure 2. Mycorrhizae of mushrooms with trees. Image from Shannon Wright
But these are only the best known of the symbiotic relationships between microorganisms and plant roots. Indeed, as the soil is full of microorganisms, many of these, including bacteria, fungi, algae, protozoa or viruses, are beneficial, symbiotic or otherwise, for the plants. And what is biotechnologically more interesting, more potential applications of these microorganisms to benefit crop plants are being found, which can be a good alternative to the use of fertilizers and pesticides.
Different microorganisms can have direct positive effects on plant nutrition as nitrogen fixation, mineralization of organic compounds, and solubilisation of elements not available to the plant (such as phosphates, K, Fe), but also indirectly positive effects, such as the production of hormones and growth factors, or protection against pathogens (García 2013).
Thus, there is a growing interest in the biological control of plant pathogens. It has been proven that some of these pathogens are inhibited by antibiotics produced by microorganisms in the rhizosphere (Raaijmakers et al 2002). Bacteria are being used (bacterization) for some years in soil or with seeds or other plant parts, with the aim of improving the growth and health of the plant.
Some of the best known and used bacteria in this sense have been Bacillus and Paenibacillus. Several species of these genera of aerobic spore bacteria are abundant in agricultural soils and can promote plant health in different ways, suppressing pathogens with antibiotic metabolites, stimulating plant defence, facilitating nutrient uptake by the plant, or promoting symbiosis with Rhizobium or with mycorrhizae (McSpadder 2004).
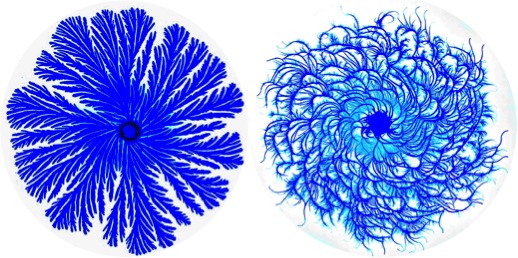
The genus Paenibacillus was reclassified from Bacillus in 1993, and includes P. polymyxa, a species N2 – fixing, which is used in agriculture and horticulture. This and other Paenibacillus species give complex and regular colonial forms in agar, even surprising (Figure 3), which vary according to environmental conditions. For this, a self-organizing and cooperative behaviour between individual bacterial cells is needed, using a system of chemical communication. This bacterial social behaviour would be an evolutive precursor of multicellular organisms.
Figure 3. Colonies of Paenibacillus dendritiformis, 6 cm diameter each, branched (left) and chiral (right) morphotypes. From Wikipedia Creative Commons.
The colonization of plant roots by these bacteria has been demonstrated, and also that they do it by forming biofilms (Figure 4). The inoculation of these bacteria to the roots promotes the growth, as shown in peppers (Figure 5). This appears to be due to the nitrogen fixing bacteria, which increases the formation of plant proteins and chlorophyll, thus increasing photosynthesis and physiological activities. And on the other hand, it has been shown that these bacteria produce siderophores, which facilitate Fe uptake by the plant (Lamsal et al 2012).
Figure 4. Colonization of Paenibacillus polymyxa and biofilm formation on roots of Arabidopsis thaliana. Adapted from Timmusk et al 2005.
Figure 5. Promoting growth effect of peppers (Capsicum annuum) by inoculation with Bacillus subtilis (AB17) and Paenibacillus polymyxa (AB15), respect the non-inoculated control. From Lamsal et al. 2012.
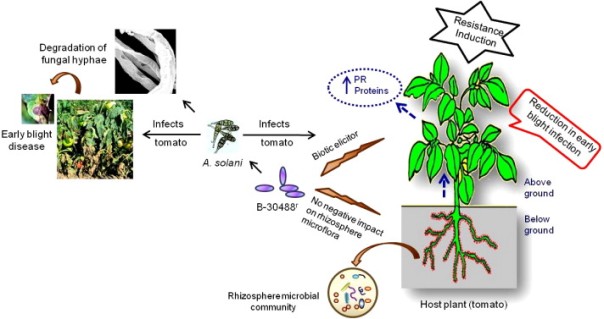
Moreover, bacteria such as Paenibacillus can be effective against plant pathogens. For example, it has been shown that a strain of P. lentimorbus (B-30488r) reduces the incidence of disease done by the fungus Alternaria solani in tomato. It has been tested (Figure 6) that after inoculating with Paenibacillus a plant infected with Alternaria, resistance to the fungus was induced in the plant. The bacteria degraded the cell walls of the fungus and also inhibited it by competition of nutrients. In addition, it was found that Paenibacillus has no negative effect on the microbial population in the rhizosphere of tomato (Khan et al 2012). These treatments are a good alternative to the use of fungicides, avoiding the environmental and health problems of these compounds.
Figure 6. Schema of the influence of Paenibacillus lentimorbus B-30488r in the interactions of tomato plant with Alternaria solani, a fungus pathogen (Khan et al 2012).
Finally, these Paenibacillus can also be useful to avoid the transmission of human pathogens such as Salmonella through the crop plants. Indeed, on the east coast of the USA a few years ago were detected outbreaks of Salmonella on tomatoes due to contamination of water. When they analyzed the microbiome present in the roots of tomatoes and these were compared with those of other places where there were no Salmonella contamination occurred, it was found that these tomatoes of the East Coast had no Paenibacillus, which were present in tomatoes of other places. With this, they decided to inoculate tomatoes with several Paenibacillus and found that Salmonella disappeared. Among the inoculated strains, one was selected as more effective, P. alvei TS -15 , for which a patent was obtained as a biocontrol agent of foodborne human pathogens (Brown et al. 2012) .
Thus, knowledge of the soil microbiota and the many forms of relationships between microorganisms and plants lead to find new strategies for using “good” microbes to prevent food safety problems of transmission of pathogens, while at the same time it can be a good ecological alternative to the massive use of pesticides.
Bibliography
Brown EW, Zheng J, Enurach A, The Government of USA (2012) Paenibacillus alvei strain TS-15 and its use in controlling pathogenic organisms. Patent WO2012166392, PCT/US2012/038584
Conniff R (2013) Super dirt. Scientific American 309, sept, 76-79.
Conniff R (2013) Tierra prodigiosa. Investigación y Ciencia 446, nov, 68-71.
García, Sady (2013) Los microorganismos del suelo y su rol en la nutrición vegetal. Simposium Perú “Manejo nutricional de cultivos de exportación”. Slideshare.net
Khan N, Mishra A, Nautiyal CS (2012) Paenibacillus lentimorbus B-30488r controls early blight disease in tomato by inducing host resistance associated gene expression and inhibiting Alternaria solani. Biological Control 62, 65-74
Lamsal K, Kim SW, Kim YS, Lee YS (2012) Application of rhizobacteria for plant growth promotion effect and biocontrol of anthracnose caused by Colletotrichum acutatum on pepper. Mycobiology 40, 244-251.
McSpadden Gardener BB (2004) Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology 94, 1252-1258
Raaijmakers JM, Vlami M, De Souza JT (2002) Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek 81, 537-547
Sánchez, Manuel. http://curiosidadesdelamicrobiologia.blogspot.com.es/2012/01/la-compania-de-transporte-paenibacillus.html
Timmusk S, Grantcharova N, Wagner EGH (2005) Paenibacillus polymyxa invades plant roots and forms biofilms. Applied and Environmental Microbiology 71, 7292-7300
Viquipèdia: http://ca.wikipedia.org/wiki/Micoriza
Wikipedia: http://en.wikipedia.org/wiki/Paenibacillus_dendritiformis
Mycobacterium tuberculosis, the pathogen bacterium that is with us since we left Africa
Tuberculosis and its agent
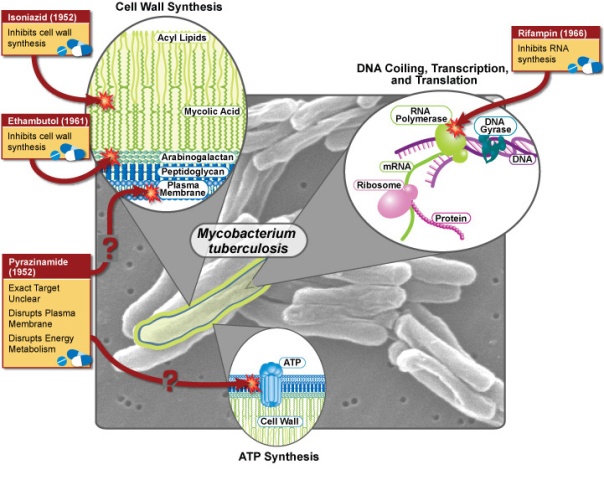
Tuberculosis is a common and often deadly infectious disease if left untreated, caused by mycobacteria, mainly Mycobacterium tuberculosis. Mycobacteria such as M. tuberculosis and M. leprae (the leprosy) are considered as gram-positive bacteria because they have glycopeptides, but they are not stained with crystal violet, because outside of glycopeptide they have a good layer of fat, called the mycolic acid (Figure 1). Precisely the prefix “myco” comes from the fungic appearance on the surface of liquid cultures of these mycobacteria.
Figure 1. Mycobacterium tuberculosis with his particular cell wall, and the more used antibiotics against this bacterium. (Figure from National Institute of Allergy and Infectious Diseases)
This mycobacterium was discovered as causing tuberculosis by the German Robert Koch (1882) and for this reason, it is also called Koch’s bacillus. It is a strict aerobic bacterium, since it needs high levels of oxygen, and therefore mostly infects the lungs. It has a generation time of about 15-20 hours, so it has a very slow growing compared to other bacteria (remember the 20 minutes in Escherichia coli). From a taxonomic point of view they are Actinobacteria, or gram-positive of high G+ C, as corynebacteria or Actinomycetales.
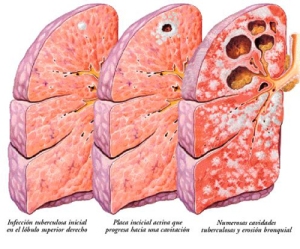
The virulence of M. tuberculosis is very complex and has many facets (Todar K.). Although apparently it produces no toxin, it has a large repertoire of physiological and structural properties that explain its virulence and pathology of tuberculosis. Once in the lungs, the bacteria are captured by alveolar macrophages, but they cannot be digested due to the structure of the bacterial cell wall and because bacteria neutralize reactive compounds from macrophages. The mycolic acid also makes the cell not permeable to lysozyme. M. tuberculosis can grow intracellularly without being affected by the immune system, and at the same time, it secretes several proteins involved in the pathogenesis. For instance, the antigen 85 complex, which binds fibronectin, facilitates the formation of tubercles in the lungs (Figure 2).
Figure 2. Progress of infection by M. tuberculosis in the lungs, with the formation of tubercles (From humanorgans.org/tuberculosis)
Tuberculosis usually attacks the lungs but can also affect many other organs. The classic symptoms of tuberculosis are a chronic cough with bloody sputum, fever and other symptoms. The diagnosis relies on radiology of the thorax, a tuberculin skin test and blood analysis, and a microscopic examination and microbiological culture of body fluids. The treatment of tuberculosis is difficult and requires long treatment with various antibiotics. The most commonly used are rifampicin and isoniazid (Figure 1). However, antibiotic resistance is a growing problem in some types of tuberculosis. Prevention is based mainly on vaccination, usually with the Bacillus Calmette-Guérin vaccine (BCG).
Tuberculosis is transmitted by air when infected people cough, sneeze or spit. One third of the world’s current population is infected with M. tuberculosis, and there is a new infected person every second. However, in most of these cases the disease is not fully developed, as the latent and asymptomatic infections are the most common. Approximately one in ten latent infections eventually it progress to active disease, which, if left untreated, kills more than half of the victims. In one year (2004), the global statistics included 14.6 million chronic active cases, 8.9 million new cases and 1.6 million deaths, mostly in developing countries. In addition, a growing number of people in the developed world are contracting tuberculosis because their immune systems are compromised by immunosuppressive drugs, substance abuse, or AIDS.
When the human tuberculosis originated ?
The origin of the population infectious diseases (such as plague, cholera, etc.), id est., those that spread easily where there are many humans together, has been associated with the Neolithic demographic transition, about 10,000 years ago, when the revolution of agriculture allowed the establishment of the first sedentary villages, with numbers of humans living together. Many of these diseases are associated with other animals, from which they are transmitted to humans. Until recently it was believed that tuberculosis was of this type, having also led the Neolithic, but in fact the majority of tuberculosis have no relationship with other animals.
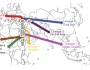
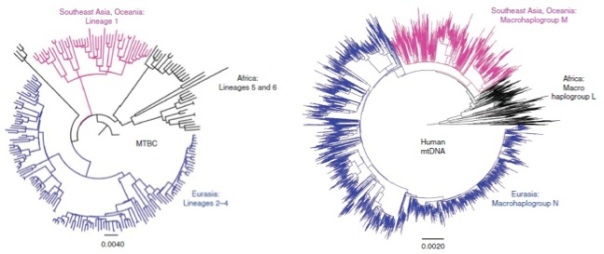
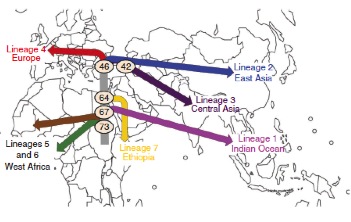
However, a recent study (Comas et al. 2013) provides data in the sense that the disease originated long before, more than 70,000 years ago, going with the modern humans out of Africa and spreading to the world. Effectively, this work of 22 co-authors from 9 countries (and the first author of which is the young Valencian Iñaki Comas) have analyzed and compared the genomes of 259 strains of the complex M. tuberculosis to see their evolutionary history. In doing so, within the 4.4 Mb genome of this species the authors have identified 34,167 SNPs polymorphic sites, i.e., sites of DNA that have some different nucleotide in function of strains, and with which they have been able to reconstruct the phylogeny of this bacterium. In this way, they have confirmed seven lineages (groups of strains) that had been suggested by other techniques. The most interesting thing is that this phylogeny of genomes of M. tuberculosis is very similar to the human mitochondrial genome phylogeny, as shown in Figure 3. Therefore, this suggests that the evolution of tuberculosis bacteria goes parallel to that of modern humans.
Figure 3. The phylogeny of genomes from 220 strains of M. tuberculosis, with the different lineages (MTBC, left), is similar to that of mitochondrial genomes (right) of 4995 humans, with their main haplogroups (from Comas et al. 2013, Fig. 1 c, d).
Based on these phylogenies and on the frequencies of mutations observed in the genomes of M. tuberculosis, the origin of the different lineages of this bacterium has been established, calculating at what time of the human Palaeolithic the different branches were appearing, from approx. 73,000 to 42,000 years (Figure 4), which, having seen the parallel with mitochondrial DNA, coincide with the proposed dates of the modern humans expansion from Africa to Eurasia.
Figure 4. Expansion of complex M. tuberculosis going out of Africa, with the origin of different lineages and the approximate data (thousands years) of the evolutionary branches (from Comas et al. 2013, Fig. 2a).
Therefore, we can conclude that tuberculosis and its bacterial agent has been a constant companion of the modern humans during their evolution and their global spread, at least in the last 70,000 years, and also that the bacterium M. tuberculosis has been able to adapt to changes in human population. In addition, in this regard last years an increase in resistant strains to multiple antibiotics has been observed. The study of these mechanisms of bacterial adaptation may help to predict future patterns of disease and to design rational strategies to fight it.
Bibliography
Comas, Iñaki, et al. (2013) Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nature Genetics 45, 1176-1182.
Todar K., Todar’s Online Textbook of Bacteriology.
Mycobacterium tuberculosis in wikipedia: http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis
Tuberculosis in viquipèdia (Catalan): http://ca.wikipedia.org/wiki/Tuberculosi.
The tropospheric bacteria of rain and snow
Nowadays there is more evidence that the bacteria found in the high troposphere (8-15 km) could influence the density of clouds and rain.
Firstly, we must remind that the troposphere is the lowest part of the atmosphere, and the 8-15 km layer is the high troposphere, near the tropopause that borders the stratosphere, above the Mount Everest. Here there are some of the highest clouds.
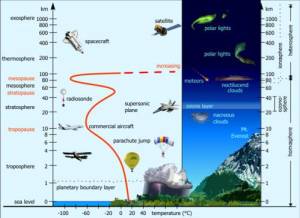 The layers of Earth’s atmosphere (www.theozonehole.com/atmosphere)
The layers of Earth’s atmosphere (www.theozonehole.com/atmosphere)
So, in a recent study (DeLeón-Rodríguez et al, 2013) it has been shown that the viable bacteria (by epifluorescence microscopy and quantitative PCR) at a 10 km altitude (samples taken above the Caribbean Sea and the Atlantic West) represent 20% of the particles with size between 0.25 and 1 mm, and bacteria are at least 10 times more abundant than fungi, with numbers of 105 per m3, with a 60% of viable cells. This suggests that bacteria are an important and underestimated fraction of microparticles of atmospheric aerosols, even at higher concentrations than lower altitudes.
The authors have analyzed the bacteria by pyrosequencing (Roche 454) the rRNA genes. They have seen that the tropospheric microbiome has a good variety of bacterial taxa that vary dynamically according to the atmospheric turbulence and in the presence of hurricanes. Some of the most abundant bacteria found are those using compounds C1-C4 (e.g., oxalic acid) present in the atmosphere, so these bacteria are metabolically active at these altitudes. This reinforces the idea of the active role of bacteria in the troposphere, and that there are not only inert spores (fungal) floating through the air.
In this sense, this metagenomic analysis also confirms the presence of bacteria that are able to catalyze the formation of ice crystals and hence the cloud condensation. This process of nucleation (ice nucleation, IN) occurs when the water molecules coalesce around a seed particle, for example dust. Depending on the temperature, these complexes can grow to become water droplets or ice, leading to the formation of rain or snow. Given that the high troposphere dust particles are scarce, it is evident the role of bacteria in this phenomenon.
One of the key roles in the nucleation of ice (IN) by bacteria is that they catalyze ice formation at temperatures close to 0°C, unlike the formation of ice nuclei by the inorganic particles, which is done at temperatures lower, below -10°C, and without any core particle the ultra-pure water freezes at -40°C.
Ice nucleation by bacteria has been reproduced in the laboratory (Christner et al, 2008) with samples of rain and snow from around the world (Canada, USA, Pyrenees, Alps and Antarctica), showing that in the samples treated with lysozyme (which hydrolyzes bacterial cell wall) or treated with heat, the IN activity was reduced almost 100% at a temperature of -5°C. Therefore, bacteria are responsible of the IN at these relatively high temperatures.
The bacteria most commonly associated with the IN activity are species associated with plants, such as Pseudomonas syringae or Xanthomonas campestris, which also often have been detected in atmospheric aerosols and clouds. P. syringae has also been found in the hail stones.
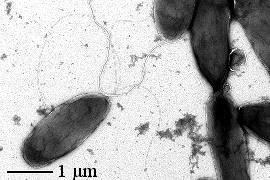 Pseudomonas syringae (www.forestry.gov.uk)
Pseudomonas syringae (www.forestry.gov.uk)
The phenomenon of IN by P. syringae was already observed in 1974 (Maki et al.) and after it has been shown (Gurian-Sherman & Lindow 1993) that IN strains of this species and others have in the outer membrane of the cell wall, as a active IN, a protein of 180 kDa, composed of repeats of a consensus octapeptide. This protein forms a planar arrangement that traps water molecules producing a mold for ice formation.
This feature makes that these bacteria are responsible for most of frost damage in plants, besides than P. syringae is pathogen of many plants at room temperature by the production of a compound (coronatin) who keeps the stomata open, causing the bacterial invasion of plant tissues (Nigel Chaffey, 2012).
Tomato leaf infected with Pseudomonas syringae (Alan Collmer, Cornell University/Wikimedia Commons)
Coming back to the frost damage, most plants can withstand up to -5°C without much damage if these bacteria are absent, but the presence of the IN protein-forming bacteria such as P. syringae in numbers of only 1000 cells by g of plant increases dramatically the damage by freezing. These damages also facilitate the penetration of bacteria and infection.
 Frozen plant (MO Plants& Maureen Gilmer)
Frozen plant (MO Plants& Maureen Gilmer)
This feature of ice nucleation by P. syringae is also utilized for the production of artificial snow. Although this can be made usually by the forced expansion of a pressurized mixture of water and air under appropriate conditions of temperature and humidity (e.g. ≤ 2°C at 20% humidity, or ≤ -2°C at 60%), snow production is favoured by the addition of nucleation agents, which can be inorganic, organic or the mentioned bacterial protein.
 Snow cannons (www.siemens.com)
Snow cannons (www.siemens.com)
Coming back to the clouds, we must remind that bacteria are far less the sole agents of nucleation forming condensation droplets resulting in rain or snow. The cloud condensation nuclei, CCN, also called cloud seeds, can be very different types of microparticles of sizes around 0.1 – 1 mm. When this aerosol of microdroplets is condensed, it forms drops of 0.02 mm in the clouds, which give falling raindrops of 2 mm.
The microparticles are mostly of natural origin such as dust, sea salt, volcanic sulphates or organic microparticles result of the oxidation of volatile compounds. Some of these may be of industrial origin, as well as soot and other particles resulting from combustion. Another important biological source of CCN is the aerosols of sulphate and methanosulphate produced from dimethyl sulphur, which is made by phytoplankton in the oceans.
Anyway, despite atmospheric microbiology is still in its infancy, as we have seen there are more and more data on the importance of bacteria and other microorganisms on bioprecipitation of rain and snow. To find out more about their role, research must go beyond the description of the abundance of microorganisms in the atmosphere, and to understand the biological, physical and chemical properties of the transport processes involved. This will require interdisciplinary approach seemingly different disciplines such as oceanography, bacterial genetics and physics of the atmosphere, for example.
 Let us imagine the bacteria (Pseudomonas syringae, photo: microbewiki.kenyon.edu) in the middle of the threatening clouds (photo: lanroca.wordpress.com)
Let us imagine the bacteria (Pseudomonas syringae, photo: microbewiki.kenyon.edu) in the middle of the threatening clouds (photo: lanroca.wordpress.com)
References
Chaffey N. (2012) COR, nice one, Mr Microbe !. AoB Blog.
Christner B. et al. (2008) Geographic, seasonal, and precipitation chemistry influence on the abundance and activity of biological ice nucleators in rain and snow. PNAS 105, 48, 18854-18859.
DeLeón-Rodríguez N. et al. (2013) Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. PNAS 110, 7, 2575-2580.
Gurian-Sherman D. & S.E. Lindow (1993) Bacterial ice nucleation: significance and molecular basis. FASEB J. 7, 14, 1338-1343.
Hardy J. (2008) The rain-making bacteria. Micro-Bytes.
http://microbialmodus.wordpress.com/tag/ice-nucleating-bacteria/
https://en.wikipedia.org/wiki/Cloud_condensation_nuclei
https://en.wikipedia.org/wiki/Snowmaking
Maki L.R. et al.(1974) Ice nucleation induced by Pseudomonas syringae. App!. Microbiol. 28, 456-460.
Morris C.E. et al. (2011) Microbiology and atmospheric processes: research challenges concerning the impact of airborne micro-organisms on the atmosphere and climate, Biogeosciences 8, 17-25
1st Conference on Research in Viticulture and Oenology in Catalonia
First of all, I apologize to friends of this blog for these last five months without any post. Sorry, I had too much work of teaching and research, and also I had to prepare this Conference that I am commenting now.
I have had the pleasure of being the organizer of this conference, along with Pep Llauradó, because I am the coordinator (and Pep is the manager) of the sub-campus Oenology of CEICS, the Campus of International Excellence Southern Catalonia.
CEICS is the strategic aggregation, driven by the Universitat Rovira i Virgili, of various institutions and structures of teaching, research, transference, and the productive sector of southern Catalonia, with the goal of becoming an international benchmark in 5 areas: Chemistry, Nutrition-Health, Tourism, Culture-Heritage and Oenology.
In 2012 I had the honor of being named coordinator of subcampus Oenology of CEICS, by the rector of the URV Xavier Grau, who is the president of CEICS.
The subcampus Oenology includes URV, Fundació URV, the technological park VITEC of Falset, IRTA, INCAVI, the cluster INNOVI, and most of catalan DO (Appelation of Origin), especially those of Tarragona, and companies and wineries, such as Freixenet and others.
One of the main objectives of CEICS is to promote the visibility of research done in the region. Therefore, since the beginning of CEICS, with the occasion of the 1st Forum held in November 2011, a conference-meeting like this was already planned, in order to share and diffuse the research done in Catalonia.
Inauguration of the Conference, with the rector of the URV Xavier Grau and the Dean of the Faculty of Oenology Joan Miquel Canals.
In Catalonia we have often the opportunity to attend various scientific conferences or technical meetings on oenology and/or viticulture. The INCAVI, the Catalan Association of Winemakers (ACE) or the cluster INNOVI, or the Facultat d’Enologia from URV often organize special courses or seminars. All of them are very interesting and necessary, and add value to the Catalan wine industry, but perhaps there had not yet been done so far a meeting of all catalan researchers in this field, in order to see all the research that is being carried out, not just in a specific focus or subject.
All in all, we considered appropriate to take the energy and push of CEICS for organizing this conference. The main objective was to publicize and disseminate the research carried out in Catalonia in Oenology and Viticulture and related issues. Before the conference, we reviewed databases of scientific publications in the last years, and we have seen that in Catalonia there are about 20 research groups whose main lines are related to the sciences of oenology and viticulture, but in addition there are 40 other groups and researchers that although his main area of research is not the viticulture and oenology, in recent years they have published several papers related to aspects of vitiviniculture, even from scientific disciplines apparently far away. All they were invited to this conference held in Tarragona (at Campus Sescelades URV), to present their most innovative research in the form of posters. I must thank them all for the good response, since there was a total of 44 posters presented.
Another implicit goal of the conference was meeting people. This conference was an opportunity to meet, discuss and explore the details of recent work with colleagues from different centers in Catalonia, maybe even meeting in person for the first time in some cases, and promote collaboration between groups.
But in addition, this Conference has not been a meeting exclusively for scientists. The research is meaningless if it has no practical application and benefits in the winemaking process, though often not immediate implementation. It binds with the main objective of the conference, as mentioned, to make visible the research done here. In this regard, some of the posters that were presented have been selected so that the authors briefly commented on the general sessions, and the selection has been done keeping in mind the interest of the research done for the wine industry.
Most of the program of the day was related to the same posters presented. So in addition to seeing and commenting particularly with authors throughout the day, there were four general sessions where experts presented them in summary. In these four sessions grouping of posters was based on their areas.
Robert Savé and Anna Puig
Lluís Tolosa taking notes of one poster.
Robert Savé (researcher of IRTA) presented and moderate the session of Viticulture. Olga Busto (professor of URV) did the same with the Oenological Chemistry and Technology session. Anna Puig (researcher of INCAVI) did the same in session of Microbiology in Enology and Viticulture, and finally the sociologist and writer Lluís Tolosa presented and moderate the session of other subjects related to the vine and wine, such as the issues of effects on health, environmental impact, socioeconomic aspects like tourism, food industry or other applications, or historical and cultural aspects and even prehistoric.
The collection of papers presented showed good international quality of research conducted in oenology and viticulture in Catalonia, and at the same time, the wide variety of topics. With the abstracts of papers a book has been edited: “1st Research Conference on Enology and Viticulture in Catalonia, Book of abstracts of the conference organized by CEICS, Tarragona June 4, 2013”, which has been published by Service of the Publications URV, with ISBN 978-84-695-7878-0.
In addition, the conference included the inaugural lesson of the internationally renowned researcher in vine genetics, José Miguel Martínez Zapater, who has an extensive curriculum research in plant genetics.
Since 1998 Dr. Zapater works on the genetics of Vitis, having achieved important results, both on varietal identification by molecular techniques and also on vine transcriptomics, with the study of gene expression in function on the conditions of climate change. Since 2010 is the director of the new Instituto de Ciencias de la Vid y el Vino (ICVV) in Logroño.
The lesson of Dr. Zapater was an excellent review of the genetic variability of Vitis and of the molecular techniques to identify different vine varieties.
We can say that the Conference, with about 70 attendees, was a success. There were researchers, students, technicians and professionals, of URV, INCAVI, IRTA, VITEC, UAB, UB, UPC, UDL, CSIC, and seven companies. In a survey that we asked them to fill up, we have concluded that most of the participants were very satisfied with the conference, its organization, its development, and express their wishdom to do it again, annualy or every two years.
The good bacteria of breast milk
Breast milk, besides being very nutritious, provides bioactive constituents that favor the development of the infant immune system and prevent diseases. From this point of view, the best known compounds are maternal immunoglobulins, immunocompetent cells and various antimicrobials. It also contains prebiotic substances, ie, several molecules such as oligosaccharides, which stimulate the growth of specific bacteria in the gut of the child.
However, other important constituents of breast milk, unsuspected until few years ago, are the bacteria. In fact, milk is not sterile, it contains microorganisms, primarily beneficial bacteria that help to establish the intestinal microbiota of the newborn, and which are the first to settle there. Although artificial milk are made to resemble the breast milk, they remain distinct and do not contain bacteria. And for this reason, the intestinal microbiota of breast-fed infants is different than those fed with artificial breast milk.
Lactobacilli (image from AJC1Flickr) and suckling baby (© Photos.com)
Just a few weeks ago was published a work ( Cabrera-Rubio et al., 2012 ) in the American Journal of Clinical Nutrition that had a good coverage in media, blogs and networks ( click here for an example), because it shows the great diversity of bacteria present in the breast milk.
Although this work done by Valencian researchers (Cavanilles Institute, University of Valencia and CSIC-IATA) with Finnish researchers is not the first study that examines this issue, this study shows that bacteria are from very diverse species.
One of the novelties of this paper is the method used, taking advantage of the latest molecular biology: they studied the microbiome in breast milk, that is, the analysis of all possible bacteria present in the samples, by DNA sequencing, without the traditional isolation of living bacteria in plates. To do so, from the aseptically collected milk, DNA is extracted and the gene fragments of bacterial 16S rRNA are amplified by PCR. These amplified genes are sequenced by pyrosequencing (454 Roche GS-FLX), the most innovative and rapid sequencing technology: a machine of this allows about 400 million base pairs (bp) of DNA in 10 hours. From the rRNA gene of each possible bacteria some 500 bp are sequenced. Thus, in this study about 120,000 sequences have been analyzed, corresponding to 2600 sequences per milk sample.
By comparing these sequences with the databases and applying statistical methods conclusions can be drawn on what taxonomic groups (genera and species) bacteria are present and in what proportion.
Predominant genera of bacteria in breast milk (Cabrera-Rubio et al., 2012)
As shown in the figure above, Cabrera et al. found in the milk of healthy mothers that the predominant genera are Leuconostoc, Weissella, Lactococcus and Staphylococcus, of which the first three are lactic acid bacteria. Although these are predominant in colostrum and milk during the first months, then other bacteria are increasing their numbers, such as Veillonella Leptotrichia (anaerobic gram-negative bacteria), which are typical commensal of the oral cavity. In total, about 1000 species have been found, that vary depending on the mother. Curiously, there are significant variations on whether delivery had been vaginal or cesarean, and on the obesity of the mother. The reasons for this are not yet clear.
And where the bacteria in breast milk come from ?
Besides the identifications made in this study of Cabrera et al. (2012) on the basis of DNA present, it has been observed by making viable counts that the total number of bacteria in breast milk is between 2·104 and 3·105 per ml (Juan Miguel Rodríguez), that is, a quantity not negligible . What is its origin?
The study of the microbiome of Cabrera et al. also concluded that the composition of different bacteria is somewhat different from that of other bacterial communities in the human body (the human bacterial niches: skin, mouth, digestive system, vagina, etc), and therefore the milk microbiome is not a particular subset of one of these niches.
The group Probilac from Universidad Complutense de Madrid, whose head is Juan Miguel Rodriguez, a friend and colleague of Red BAL (Spanish network of lactic acid bacteria) is working in this area for years (ex: Martin et al 2003 , Martin et al 2004).
As discussed in a recent review published by this group (Fernández et al 2012), the bacteria present in the breast milk would come from three possible sources (figure below): skin bacteria from the same breast, the oral cavity of the infant, and the most surprising, commensal bacteria of the maternal gut that pass to milk by the entero-mammary pathway.
Potential sources of bacteria present in human colostrum and milk, including the transit of intestinal commensal bacteria to the milk by the entero-mammary pathway (Fernández et al., 2012). DC: dendritic cells.
Indeed, several studies had shown that dendritic cells cross the intestinal epithelium (between enterocytes) and may take commensal bacteria of the gut lumen, incorporating them by endocytosis, but keeping them alive. See details in the following diagram.
Dendritic cell capturing gut bacteria (Scheme of J.M. Rodríguez, group Probilac, Univ. Complutense de Madrid).
These dendritic cells travel through the circulatory system, reaching the mammary glands, where it seems that include bacteria to milk. This is the the entero-mammary pathway.
In this breast microbiota, bacteria from breast skin and from oral cavity of the child also would be incorporated. Some of these bacteria the child’s oral cavity are actually related to those of its gastrointestinal tract. As the first bacteria inhabiting this tract are those of the vaginal microbiota during birth (and intestinal if delivery is cesarean), this would explain the phylogeny of certain bacteria in the milk of these microbiota.
In summary, we see as the “good” bacteria (lactic acid bacteria, but also bifidobacteria and other) from maternal gut, by different ways, arrive to breast milk, and the reach the child’s gut, developing there the child’s microbiota, and helping to complete the neonatal immune system.
Bibliography
Cabrera-Rubio R, MC Collado, K Laitinen, S Salminen, E Isolauri, A Mira (2012) The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. American J Clinical Nutrition 96, 544–51
Grupo Probilac (Juan Miguel Rodríguez Gómez) Microbiota de la leche humana en condiciones fisiológicas: http://www.ucm.es/info/probilac/microbiota2.htm, Departamento de Nutrición, Bromatología y Tecnología de los Alimentos, Facultad de Veterinaria, Universidad Complutense de Madrid
Fernández L, S Langa, V Martín, A Maldonado, E Jiménez, R Martín, JM Rodríguez (2012) The human milk microbiota: Origin and potential roles in health and disease. Pharmacological Research http://dx.doi.org/10.1016/j.phrs.2012.09.001
Hunt KM JA Foster, LJ Forney, UME Schütte, DL Beck, Z Abdo, LK Fox, JE Williams, MK McGuire, MA McGuire (2011) Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 6:e21313.
Martín R, S Langa, C Revriego, E Jiménez, ML Marín, J Xaus, L Fernández, JM Rodríguez (2003) Human milk is a source of lactic acid bacteria for the infant gut. J Ped. 143, 754-758.
Martín R, S Langa, C reviriego, E Jiménez, ML Marín, M Olivares, J Boza, J Jiménez, L fernández, J Xaus, JM Rodríguez (2004) The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci Technol 15:121–7.
Other references
Adlerberth I (2006) Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle. Pediatric Res 59, 96-101.
Albesharata R et al (2011) Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst App Microb 34, 148–155
Domínguez-Bello MG et al (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA;107:11971–5.
Huurre A et al (2008) Mode of delivery—effects on gut microbiota and humoral immunity. Neonatology;93:236–40
LeBouder E et al (2006) Modulation of neonatal microbial recognition: TLRmediated innate immune responses are specifically and differentially modulated by human milk. J Immunol;176:3742–52.
Martín R et al (2009) Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol 75:965–9.
Pérez PF et al (2007) Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119: 724–732.
Rescigno M et al (2001) Dendritic cells shuttle microbes across gut epithelial monolayers. Immunobiology 204:572–81.
Stockinger S et al (2001) Establishment of intestinal homeostasis during the neonatal period. Cell Mol Life Sci;68: 3699–712.